
Electronic health records (EHR) represent the basis for the automation of a medical organization. Recently, EHRs have been widely implemented all over the world.
According to some reports, about 95% of US hospitals have implemented a wide variety of EHR management systems. In Russia many medical organizations have begun to use EHR, thanks to the implementation of the Uniform State Health Information System (USHIS) in 2011-2018, and then to the subsequent federal project on creation of a unified digital circuit in the healthcare based on the USHIS.
Some advanced regions and individual healthcare organizations have been working with such systems for 5 or more years, having accumulated truly huge databases with a variety of protocols for instrumental and laboratory examinations, medical examinations, etc.
In general, in the healthcare digital transformation industry, it is believed that EHRs can rarely be used for machine processing, especially for creating datasets based on them for subsequent machine learning.
The main reason is the wide spread of unstructured medical records, especially medical examinations. Indeed, today most EHR systems focus on the convenience of users and the speed of entry, which is most often done through different templates and simplified text forms like mini-text editors. The reason is that if a doctor is forced to enter information through rigid formalized input forms, even though such information will be more structured (and, in part, of better quality), it will take significantly more time to enter it.
According to the observation of our experts, sometimes this method requires 2 to 3 times more time than a quick correction of an ordinary text template, loaded from a reference book of text templates.

We believe that this situation will persist for quite a long time. Therefore, unstructured medical examination protocols will prevail over detailed machine-readable formalized clinical protocols. Hence the idea is not to deny unstructured health records, but to learn to work with them.
Specialized services are created for such tasks. One such solution is Amazon Comprehend Medical, which allows you to extract meaningful information (complaints, diagnosis, prescribed drugs and dosage, test results, etc.) from unstructured health records.
At the same time, no machine learning knowledge is required from the user as Comprehend Medical is provided as a service through Amazon's Integration APIs. This product is aimed at healthcare providers, insurers and researchers, as well as medical, biotech and pharmaceutical companies, which will quickly implement decision support systems and improve patient health data management https://aws.amazon.com/ru/blogs/machine-learning/introducing-medical-language-processing-with-amazon-comprehend-medical/.
This approach has recently been confirmed in serious scientific literature. For example, at the end of 2019, JAMIA published a study that showed that data obtained from unstructured EHRs is a more accurate source of information for predicting CHD than structured data. This further reinforces our conviction that extracting features from unstructured EHR has significant potential, incl. constructing large datasets for machine learning. Moreover, AI development allows not only to extract features from records, but demonstrates that working with unstructured records can give AI models even higher accuracy than if the features were formalized https://www.healthcareitnews.com/news/unstructured-ehr-data-more-useful-predictive-analytics-study-shows.
In modern EHR, up to 70% of information is written in natural language. Free text is useful for expressing clinical concepts and events such as diagnostics, symptoms, and interventions. Doctors record patient complaints, symptoms, and prescribed medication in their notes in unstructured format. Many important observations remain unregistered in the fields of the protocol forms, very often you can see the doctor's comments in the form of free text stored next to the empty fields of the forms.
In order for the Webiomed system to extract structured features from medical records, we have created a special service, Webiomed.NLP.
In order to develop it, we build machine learning models using Natural language processing (NLP) methods. It allows our system to extract clinically significant unlabeled information from ordinary textual medical protocols, which is then used for machine analysis of requests from medical information systems received by Webiomed.
With the help of NLP we extract symptoms from complaints, blood pressure data, patient height and weight from unstructured objective data, laboratory indicators from discharge lists and much more. We use Webiomed.NLP to enrich the accumulated raw data and use it to create our own labeled datasets, which we need for subsequent machine learning and predictive models construction.
Methods
For medical text records analysis special methods and Named Entity Recognition (NER) are used. These methods are applied for labeling and classification of tagged entities in text (classes). The number of classes is known in advance. Classes can be names of diseases, facts of a fatal case or hospitalization of a patient (yes or ni), various quantitative and qualitative parameters, signs and cases, etc.
Most of NER classifiers are based on the CRF (Conditional random field) algorithm, which belongs to the class of hidden Markov models https://nlp.stanford.edu/software/CRF-NER.shtml.
To create NER classification models, we use the open source natural language processing (NLP) framework SpaCy. It is written in Python and performs tokenization, part-of-speech (PoS) marking, and dependency parsing.
The study by Choi et al. showed that Spacy is the fastest NLP analyzer available, providing high accuracy of feature extraction. In the paper ‘It Depends: Dependency Parser Comparison Using A Web-based Evaluation Tool’ 10 different off-the-shelf feature extraction systems were evaluated for accuracy and extraction speed. As a result, SpaCy showed the best overall speed, maintaining comparable accuracy from 85% to 90%.
SpaCy models are convolutional neural networks (CNNs) that allow you to make predictions based on the examples presented during training. To train the Spacy model, you need tagged texts of named entities that the model should predict. More information can be found here: https://spacy.io/.
In addition to Spacy, the following packages and libraries are used when developing machine learning models: Keras, OpenNN, PaddlePaddle, PyTorch, TensorFlow, Theano, Torch.
Model training
In order to create models, we accumulate a variety of Russian data from electronic health records with various diseases: cardiovascular diseases, pneumonia, gastritis, ARVI, COPD, etc. NER models are trained based on depersonalized data from the EHR. Mostly these are medical records of patients aged 30 to 80 years, which contain unstructured text from the records made during the initial examination, including objective data (height, weight, blood pressure, respiratory rate), patient complaints (fever, shortness of breath, cough, heartburn), as well as signs of laboratory data (glucose, cholesterol, creatinine, urea). To extract each medical text during training, about 200-500 labeled texts with different spellings by doctors are used.
Model metrics
For assessing NER tasks the following metrics are used:
- Precision
- Recall
- F1 - harmonic mean of the model’s precision and recall
Due to the successful application of this approach in extracting any information, it was agreed that SOTA NER (best metric value) is considered when F1 is greater than 0.9. In our models, the F-measure is about 0.986.
Developed models of feature extraction from medical records
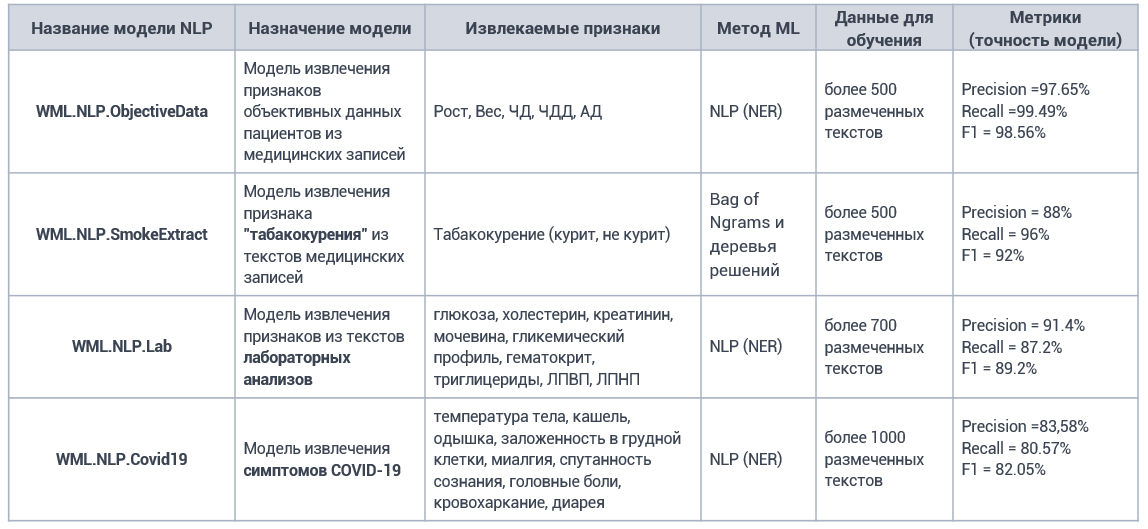
An example of Webiomed service extracting features from medical records
Consider one example of a medical record (in Russian):
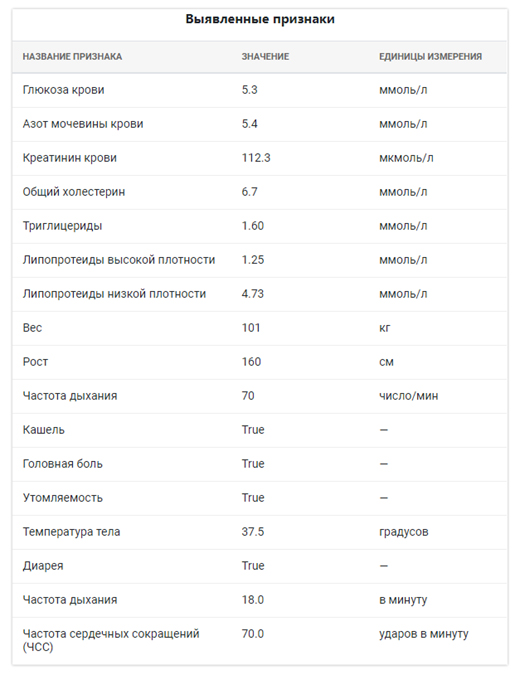
“Жалобы на малопродуктивный кашель, насморк, першение в горле, головные боли, общую слабость, Т 37,5. Около 1 недели назад были боли в животе, однократная рвота, вызвыали СМП. На момент вызова болей нет, беспокоят ежедневный жидкий стул, мацерация ануса. OBJECTIVE: Вес-101, рост- 160 см., АД 140/100, Курит 5 лет. Кожа чистая. Периферические лимфоузлы не увеличены. Щитовидная железа 0 ст. Дыхание везикулярное, без хрипов. ЧД- 18. ЧСС 70 в мин., ритмичный. Б/х крови 27.09.17г.: Общ.белок 72.3 г/л, Билирубин общ. 12.4 мкмоль/л, пр. - 2.4 мкмоль/л, АЛТ 23.8 Е/л, АСТ 34.0 Е/л, Глюкоза крови 5.3 ммоль/л, Мочевина 5.4 ммоль/л, Креатинин 112.3 мкмоль/л, Холестерин 6.7 ммоль/л, Триглицериды 1.60 ммоль/л, РФ - отр., СРБ 36 мг/л, ЛПВП 1.25 ммоль/л, ЛПНП 4.73 ммоль/л, ПТИ 103.1%. МНО 0.96."
The results of feature extraction:
Conclusions:
- The models for extracting features from medical text records by NLP methods developed by the company have shown high accuracy in processing unstructured EHR. At the moment, they are undergoing comprehensive testing on pilot-project data from medical organizations in order to identify predictors of disease development.
- It is planned to significantly expand the number of extracted features by NLP models to further develop the predictive capabilities of our system for various diseases and risk factors of patients.
- The feature extraction method allows using a very large amount of EHR data for machine learning. Hundreds of thousands of records can be used, which is almost impossible to implement in clinical studies, such as the Framingham Heart Study (about 10 thousand patients were examined over a long period).
- The Webiomed.NLP service can be integrated into any medical information system in order to increase its functionality in interpreting medical records in EHR.
The results of our work were presented in the publication: